|
Application Note #1: Flow Cytometry analysis
Updated 09/20/00 for PhiPhiLux-G1D2
New protocol in blue!
Substrate composition and fluorescence characteristics:
Each substrate molecule contains a peptide homodoubly
labeled with a fluorophore. The cleaved substrate has the following fluorescence peak
characteristics:
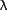 ex=
505 nm and ex=
505 nm and 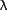 em= 530
nm. (The fluorescence of the precleaved fluorogenic protease substrate is not
completely quenched.) em= 530
nm. (The fluorescence of the precleaved fluorogenic protease substrate is not
completely quenched.)
The main peptide amino acid sequence is GDEVDGI.
Components: The caspase substrate reagent kit (cat. #A304R1G-3)
contains three vials (each vial contains at least 500 ul) plus one additional substrate
vial containing at least 750 ul of 10 uM substrate in RPMI 1640 medium with 25 mM HEPES. Additionally, 1 bottle (60 ml) of flow cytometry
dilution buffer is included. The caspase substrate reagent kit (cat. #A304R1G-6) contains
six vials (500 ul each) plus two additional substrate vials (750 ul each) of 10 uM
substrate in RPMI 1640 medium with 25 mM HEPES and 2 bottles
of the flow cytometry dilution buffer.
Use the contents of the 750 ul substrate vial(s) either to test your
own experimental protocol or to increase the substrate solution volume per sample from 50
ul to 75 ul. The latter will reduce the risk of diluting the substrate concentration from
residual medium. Hence, both the 30 and 60 test kit sizes contain enough substrate
for 45 and 90 tests if only 50 ul substrate solution per sample is used.
Possible additional reagent needed: 250 ul of fetal calf serum (FCS).
Before storing: Please
centrifuge the reagent-containing vials lightly to remove any liquid from caps, then store
at -10 to -20 oC.
Upon thawing: Shake
until the solution appears homogeneous.
Incubation conditions:
Do not fix cells that have been previously
incubated with the substrate for antibody labeling or labeling with other reagents. Remove
all steps involving fixation or permeabilization from your protocol.
1. Treat target cells with chosen apoptosis-inducing
reagent and/or inhibitor. (See D below.)
2. Aliquot cells into 1.5 to 2.0 ml microcentrifuge tubes, then
centrifuge and remove all of the culture medium in order to
minimize substrate dilution. Use a gentle vacuum suction
equipped with a capillary tip to completely remove the medium from microfuge tubes with
the centrifuged cell pellets.
3. To each of the centrifuged cell pellets, add 50 to 75 ul of
10 uM substrate solution (and add 5 ul of FCS, if 10% FCS is appropriate). The cell number
in these solutions should be between 0.5 and 1 million per sample (See A and B
below). Mix suspension containing cells and substrate by flicking tubes with finger tips. Do
not vortex tubes containing cells. Apoptotic cells can
be “fragile’”.
4. Incubate tubes at 370 C
for 60 minutes before flow cytometric analysis. (See C below.)
5. Avoid direct light to substrate and exposure to extremes of pH. Keep
substrate at physiological pH.
6. Recommended flow cytometer setting: FL1 channel of a BD instrument
with excitation at 488 nm. Use Fl3 for PI.
Sample preparation for flow cytometry:
7. Wash cells once by adding 1 ml of ice cold
flow cytometry dilution buffer, centrifuging, and removing all of buffer. Loosen the cells
pellets by flicking the tubes with finger tips and then resuspending the loosen pellets in
1 ml of fresh dilution buffer. Do not vortex tubes containing cells, but rather
flick tubes with finger tips to resuspend cells. Apoptotic
cells can be “fragile”. (See D.)
8. Keep cell suspension on ice until analysis by flow cytometry.
9. All samples should be analyzed within 60 to 90 minutes after the end
of the 37 oC incubation.
10. After collecting data by the preceding procedure, one can delete
PI(+) cells if a drop of a 5-10 ug/ml propidium iodide (PI) solution is added and samples
are rerun on the flow cytometer (final PI
concentration of @200ng/ml) (See D). Reanalysis should be within
5 minutes of PI addition. Alternatively, if
an additional 2.0 ml of flow cytometry dilution buffer (or PBS) is added to each sample,
then cells that would be PI(+) will shift to a lower intensity range due to the leakage of
intact PhiPhiLux substrate. Thus, the fluorescence of dead cells, i.e., those that
would be PI(+), may show lower fluorescence intensity in the FL1 channel than will
apoptotic PI(-) or non-apoptotic cells. (See D below.)
Useful Hints & Warning:
A. The cell density during incubation with the substrate can be as high
as 0.5 to 1 million cells per sample. It is recommended that a control sample with the
cells taken directly from a culture with cells in log phase be included in the assay to
test whether there is any high cell density-induced apoptosis.
B. In certain settings, one may be able to use the substrate at a
concentration lower than 9 uM [addition of FCS to a final concentration of 10% v/v would
lower the substrate concentration to 9 uM.]. However, the recommended final
substrate concentration should not be less than 9 uM.
C. Viable cell uptake of the substrate reaches a near maximum
by 15 to 20 minutes at 37 oC. as stated above, the incubation time may vary
with cell type.
D. We recommend that in order to identify PI(-)
apoptotic and uninduced cell populations, one analyze samples, first, without PI addition
and then the same samples after PI addition. When one compares the dot plots of FL1 vs
FL3 (or FL2) of these two samples, one can easily identify the PI(-) cell population as
those cells that remain in the same location in the FL1 vs FL3 [FL2] dot plot after
PI has been added. Place a gate around the PI(-) population and plot this gated FL1
histogram. This histogram should show both uninduced and apoptotic cells. One should avoid
use of a threshold-based (orthogonal) gate for PI(+) cells, i.e., designation of
cells above a certain FL2 or FL3 channel number as PI(+), since some very bright PI(-)
apoptotic cells may appear in a region defined as PI(+). Be sure that the sample
dilution volume is the same for all samples.
If one observes the appearance of a very low fluorescence intensity
population, i.e., lower than the uninduced cell population, then more than likely
the samples have been overtreated with inducing agent or conditions that over induce the
sample. In order to see the brighter apoptotic cell populations in histograms, these
apoptotic cells must retain their membrane integrity. Pleae note that caspase-cleaved
substrate fragments which generate positive signal in the PI(-) apoptotic cells are
retained since the fragments diffuse through intact membranes significantly slower than
the intact substrate. This differential diffusion rate will be lost when the cells loose
their membrane integrity. When membrane integrity is lost, the cleaved substrate fragments
leak out of such cells and these cells appear dark.
Generally, cells in samples with a high percentage of PI(+) or TUNEL
assay positive cells will be in late apoptosis and/or necrosis. Use of PhiPhiLux is
optimal with samples containing a low percentage of PI(+) cells. In most cases we
recommend lowering the inducing agent concentration rather than simply looking for an
earlier time point. Appropriate time points for PhiPhiLux analysis of apoptotic cells
should be such that a large percentage of viable PI(-) cells is present and the samples
contain both uninduced cells plus caspase(+)/PI(-) cells. Examination of cells under a
fluorescence microscope may assist in determining exact apoptosis inducing conditions.
Application Note #2 :
Fluorescence Microscopy
Updated 09/20/00
Incubation conditions:
Do not fix cells that have been previously
incubated with the substrate for antibody labeling or labeling with other reagents. Remove
all steps involving fixation or permeabilization.
1. Treat target cells with chosen apoptosis-inducing
reagent and/or inhibitor. (See D of Application #1: Flow Cytometry Application.)
2. (a) For suspension cells, aliquot into 1.5 to 2.0 ml microcentrifuge
tubes, then centrifuge and remove as much the culture medium as possible in order to
minimize the substrate dilution. (b) For adherent cells, remove all culture medium.
3. (a) For suspension cells, to each of the centrifuged cell pellets,
add 50 to 75 ul of 10 uM substrate solution (and add 5 ul of FCS, if 10% FCS is
appropriate). The cell number in these solutions should be between 0.5 and 1
million per sample (See A and B in Application #1: Flow Cytometry Application). Mix
suspension containing cells and substrate by flicking tubes with finger tips. Do not
vortex tubes containing cells. Apoptotic cells are
“fragile”. (b) For adherent cells, add enough substrate solution to completely
cover the monolayer or individual adherent cells. With either configuration be sure to
remove all medium before addition of substrate-containing solution to minimize dilution of
substrate.
4. Incubate suspension of cell samples in tubes
or adherent cells at 37 oC for 30 to 60 minutes. (For adherent cells we
recommend a 35 mm cell culture dish with a glass coverslip attached to the bottom side).
Exact incubation times are cell type and inducer specific.
5. Avoid direct light to substrate and exposure to extremes of pH.
Keep substrate at physiological pH.
6. Immediately following incubation with PhiPhiLux: (a) for suspension
cells dilute with 1.5 ml of the physiological buffer, e.g.,
PBS at 4 oC, centrifuge, and replace the supernatant with 1.5 ml of fresh
buffer (at 4 oC). Repeat this cell washing process two to four more times
depending on your fluorescence microscope’s lamp power and detection capability.
Check cells after each wash under fluorescence microscope to see if the background
fluorescence is dark enough to distinguish between untreated control and treated cells.
(b) for adherent cells remove PhiPhiLux-containing medium and wash gently with buffer.
Perform a similar number of cell washing cycles as suspension cell samples. After each
cycle make observation under fluorescence microscope to evaluate the background
fluorescence level. Take special care in washing adherent cells since apoptotic cells
are generally more easily detached from the plate than nonapoptotic cells.
7. Recommended microscopy settings are fluorescein filters.
Please contact us for ordering
information for sterile 35 mm culture dishes with glass centers for use with inverted
fluorescence microscopes.
References (updated 05/31/00):
On Exciton theory used in fluorogenic protease substrate
design:
1. B.Z. Packard, D.D. Toptygin, A. Komoriya, and L. Brand
Profluorescenct protease substrates: intramolcular dimers described by the exciton model.
Proc. Natl. Acad. Sci. (USA) 93: 11640-11645 (1996).
2. B.Z. Packard, D.D. Toptygin, A. Komoriya, and L. Brand. The design
of fluorogenic protease substrates guided by exciton theory. Meth. Enzym. 278:
15-28 (1997).
3. B.Z.Packard, A. Komoriya, D.D.Toptygin, and L. Brand. Structural
characteristics of fluorophores which form intramolecular H-type dimers in a protease
substrate. J. Phys. Chem. B 101: 5070-5074 (1997).
4. B.Z. Packard, D.D. Toptygin, A. Komoriya, and L. Brand.
Characterization of fluorescence quenching in bifluorophoric protease substrates. Biophys.
Chem. 67: 167-176 (1997)
5. B. Z. Packard, D.D. Toptygin, A. Komoriya, and L. Brand.
Intramolecular resonance dipole-dipole interactions in a protease substrate. J. Phys.
Chem. B 102: 752-758 (1998).
6. B.Z. Packard, A. Komoriya, V. Nanda, and L. Brand. Intramolecular
excitonic dimers in protease substrates: modification of the backbone moiety to probe the
H-dimer structure. J. Phys. Chem. B 102: 1820-1827 (1998).
7. A. Komoriya, B. Z. Packard, M. J. Brown, M.-L. Wu, and P. A.
Henkart. Assessment of Caspase Activities in Intact Apoptotic Thymocytes Using
Cell-permeable Fluorogenic Caspase Substrates. J. Exp. Med. 191:1819-1828 (2000)
On Caspase-3 nomenclature:
8. E.S. Alnemri et al. Human ICE/CED-3 Protease Nomenclature. Cell 87:
171 (1996)
On DEVD-based substrate and inhibitor characteristics:
9. A. Sarin, M.-L. Wu, and P.A. Henkart. Different Interleukin-1b
Converting Enzyme (ICE) family protease requirements for the apoptotic death of
T-lymphocytes triggered by diverse stimuli. J. Exp. Med. 184: 2445-2450 (1996)
10. D.W. Nicholson et. al. Identification and inhibition of the
ICE/CED-3 protease necessary for mammalian apoptosis. Nature 376: 37-43 (1995).
11. N.A. Thornberry, T.A. Rano, S.Roy, J.P. Vaillancourt, K.T. Chapman,
and D.W. Nicholson. A Combinatorial Approach Defines Specificities of Members of the
Caspase Family and Granzyme B Functional Relationships established for Key Mediators of
Apoptosis. J. Biol. Chem. 272:17907-17911 (1997)
Intracellular Caspase-3-like activities determination using
PhiPhiLux:
12. H. Hirata, A. Takahashi, S. Kobayashi, S. Yonehara, H. Sawai, T.
Okasaki, K. Yamamoto, and M. Sasada. Caspases are activated in a branched protease cascade
and control distinct downstream processes in Fas-induced apoptosis. J. Exp. Med. 187:
587-600 (1998)
13. J. M. Zapata, R. Takahashi, G.S.Salvesen and J.C. Reed. Granzyme
release and caspase activation in activated human T-lymphocytes. J. Biol. Chem. 273:
6916-6920 (1998).
14. R.M. Siegel, D.A. Martin, L.Zheng, S.Y. Ng, J. Cohen, and M.J.
Lenardo. Death-effector filaments: novel cytoplasmic structures that recruite caspases and
trigger apoptosis. J. Cell Biol. 141: 1243-1253 (1998).
15. L. Guedez, W.G. Stetler-Stevenson, L. Wolff, J. Wang, P. Fukushima,
A. Mansoor, M. Stetler-Stevenson. In vitro suppression of programmed cell death of B cells
by tissue inhibitor of metalloproteinases-1. J. Clin. Invest. 102: 2002-10 (1998).
16. P.G. Ekert, J. Silke and D.L. Vaux. Inhibition of apoptosis and
clonogenic survival of cells expressing crmA variants: optimal caspase substrates are not
necessarily optimal inhibitors. EMBO J. 18: 330-338 (1999).
17. D.M. Finucane, E. Bossy-Wetzel, N.J. Waterhouse, T.G. Cotter and
D.R. Green. Bax-induced caspase activation and apoptosis via cytochrome c release
from mitochondria is inhibitable by Bcl-xL. J. Biol. Chem. 274: 2225-2233 (1999).
18. G.I. Perez, X-J. Tao and J.L. Tilly. Fragmentation and death
(a.k.a. apoptosis) of ovulated oocytes. Molecular Human Reproduction 5: 414-420
(1999).
19. R. Robles, X-J. Tao, A.M. Trbovich, D.V. Maravei, R. Nahum, G.L.
Perez, K.L. Tilly and J.L. Tilly. Localization, regulation and possible consequence of
apoptotic protease activating factor-1(Apaf-1) expression in granulosa cells of the mouse
ovary. Endocrinology 140: 2641-2644 (1999).
20. H. Kanuka, S. Hisahara, K. Sawamoto, S. Shoji, H. Okano and M.
Miura. Proapoptotic activity of Caenorhabditis elegans CED-4 protein in Drosophila:
Implicated mechanisms for caspase activation. Proc. Natl. Acad. Sci. 96: 145-150
(1999).
21. B. Huppertz, H.-G. Frank, and P. Kaufmann. The apoptosis cascade --
morphological and immunohistochemical methods for its visualization. Anat. Embryol. 200:
1-18 (1999).
22. B. Huppertz, H.-G. Frank, F. Reister, J. Kingdom, H. Korr, and P.
Kaufmann. Apoptosis Cascade Progressess during Turnover of Human Trophoblast: Analysis of
Villous Cytotrophoblast and Syncytial Fragments in Vitro. Laboratory Investigation 79:1687-1702
(1999).
23. K.J. Harvey, D. Lukovic, and D.S. Ucker.
Caspase-dependent Cdk activity is a requisite effector of apoptotic death events. J. Cell
Biol. 148:59-72 (2000).
24. A. Komoriya, B.Z. Packard, M.J. Brown, M.-L. Wu, and
P.A. Henkart. Assessment of Caspase Activities in Intact Apoptotic Thymocytes Using
Cell-permeable Fluorogenic Caspase Substrates. J. Exp. Med. 191:1819-1828 (2000).
Updated 9/20/00.
Copyright © 1998-2001 OncoImmunin Inc. All rights reserved.
|